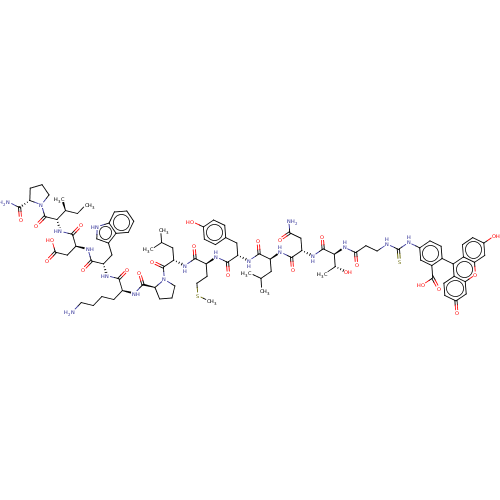
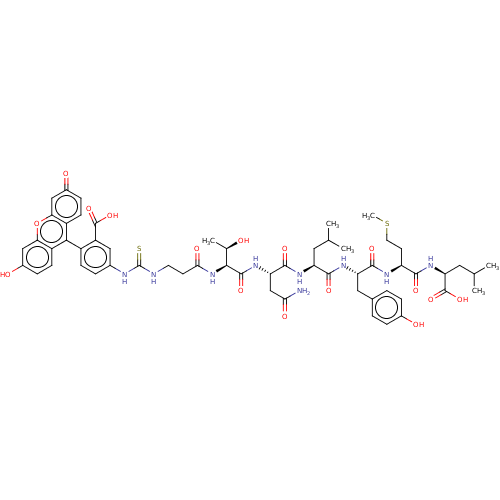
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 600nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair
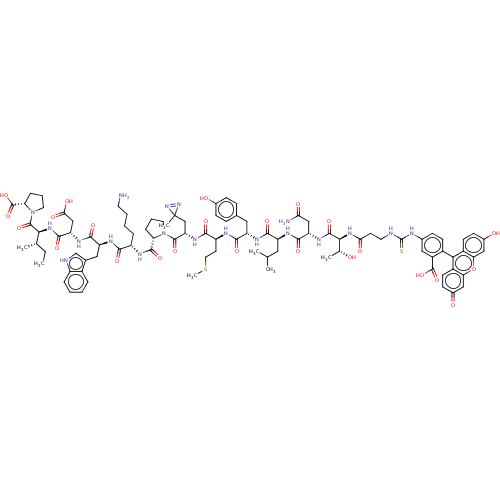
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 800nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair
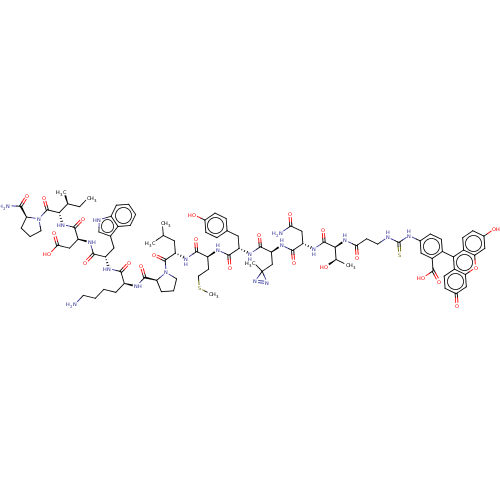
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 1.10E+3nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair
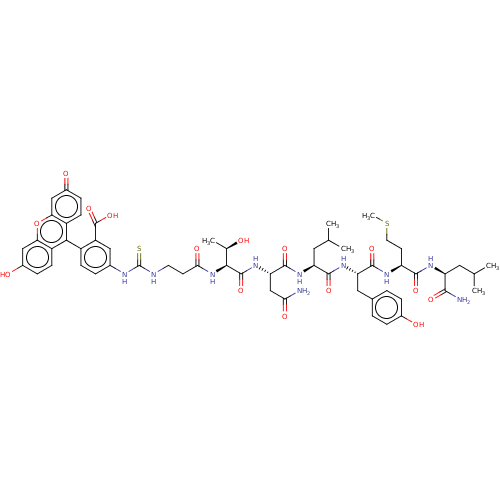
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 2.10E+3nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 2.20E+4nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair
TargetUDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase(Escherichia coli (Enterobacteria))
University of Michigan
University of Michigan
Affinity DataKd: 3.60E+4nMpH: 8.0Assay Description:The fluorescence polarization assay was performed as previously described [Jenkins et al., ACS Chem. Biol., 7:1170-1177]. In 384-well black Costar pl...More data for this Ligand-Target Pair